Match-a-Yak
You must be a member, and logged in, to use this.
Match-a-Yak is a tool meant to help make pairing decisions for breeding. The North American yak gene-pool is very narrow, and pedigrees are frequently very short. Under these circumstances it is possible to unknowingly breed closely related yaks.
This tool can be used to roughly forecast the Coefficient of Inbreeding (COI) that would result from breeding a bull with various females in a herd. Using this COI tool can offer a reference in regard to which females to place with specific bulls.
Pedigree information should still be used and considered. This computer program is not (yet) meant to replace pedigree information. The COI result can serve as a valuable supplement. If you have little or no pedigree information, the COI is the only tool available to scientifically guide your breeding decisions.
The USYAKSID (aka registration number) is needed to use the Match-a-Yak process. Enter the USYAKSID in the appropriate field and push the <Run Match> button. The program will forecast the COI of a calf produced from your trial pairing.
Information on COI from the USYAKS registry page:
To interpret the number displayed in a COI result, from a mathematical standpoint, every COI number is between -1.0 and +1.0. In actual practice, the observed COIs vary between about +0.45 (greatest in-breeding) and -0.45 (greatest out-crossing). The average COI is zero. A COI in the positive range means a yak is more in-bred than average. A COI in the negative range indicates less in-breeding than average. About 50% of registered yaks have a COI between -0.10 and +0.10.
The USYAKS genetic test examines a very narrow slice of a yak’s total DNA, so the actual COI of an individual calf can vary from the predicted COI.
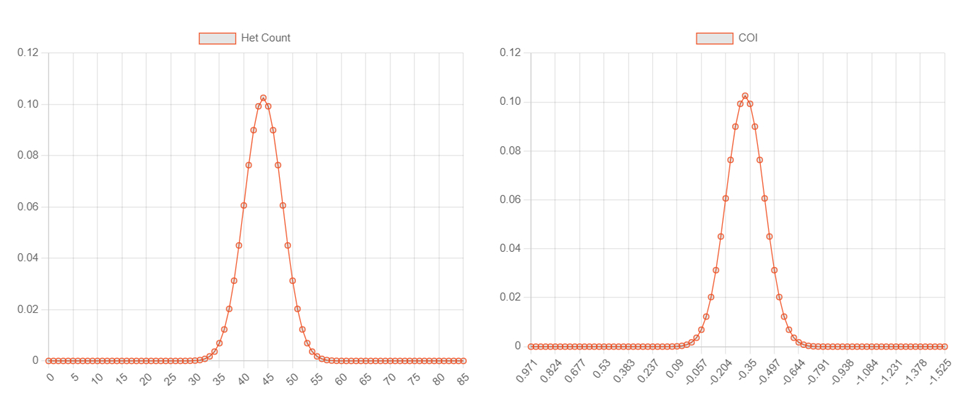
The computer output of COI result is also displayed in a bell curve given “likelihoods” of COI, mitochondrial DNA (mtDNA) and predicted degree of cattle gene introgression.
The strength of this COI result will continue to develop and improve as USYAKS increases the capacity to do more in-depth genetic testing in yak. As a yak’s DNA is studied in greater depth, the preciseness of COI forecasts will improve, eventually resulting in a narrowing of the bell-shaped curves.
USYAKS has a Science-Based-Registry and is committed to advancing the science that enables the construction, research and development of tools like this one.
Please use this free Match-a-Yak tool and send any comments or suggestions regarding your experience with it to President@USYAKS.org . The association is committed to improving and developing tools for yak owners as the science and research of yak DNA continues.
Hypothetical example of output obtained by Match-a-Yak using bull 09B78 with dam 09C08:
Predicted hetsites for calf of this pair: 40.5
Number of useable sites: 85
Expected Heterozygote proportion: 0.39608279
Predicted calf COI: -0.20295705914234285
Expected number of cattle alleles: 0
Expected mitochondrial DNA type: G – Cattle
| SNP # | HSY Yoda | HSY Amber | Probability of Hetsite | Probability of Homsite |
|---|---|---|---|---|
| 1 | G/T | G/G | 0.5 | 0.5 |
| 2 | C/T | C/T | 0.5 | 0.5 |
| 3 | T/T | G/T | 0.5 | 0.5 |
| 4 | C/T | C/C | 0.5 | 0.5 |
| 5 | A/T | A/T | 0.5 | 0.5 |
| 6 | G/T | G/T | 0.5 | 0.5 |
| 7 | T/T | T/T | 0 | 1 |
| 8 | A/G | A/A | 0.5 | 0.5 |
| 9 | skip | skip | ||
| 10 | C/C | C/G | 0.5 | 0.5 |
| 11 | C/T | C/T | 0.5 | 0.5 |
| 12 | G/G | C/G | 0.5 | 0.5 |
| 13 | G/G | G/G | 0 | 1 |
| 14 | C/C | T/T | 1 | 0 |
| 15 | C/C | T/T | 1 | 0 |
| 16 | A/T | A/T | 0.5 | 0.5 |
| 17 | C/T | C/C | 0.5 | 0.5 |
| 18 | G/G | G/G | 0 | 1 |
| 19 | C/C | C/C | 0 | 1 |
| 20 | A/T | A/T | 0.5 | 0.5 |
| 21 | C/C | C/T | 0.5 | 0.5 |
| 22 | A/A | A/A | 0 | 1 |
| 23 | C/T | C/T | 0.5 | 0.5 |
| 24 | C/C | G/G | 1 | 0 |
| 25 | C/T | C/T | 0.5 | 0.5 |
| 26 | G/G | G/T | 0.5 | 0.5 |
| 27 | A/A | G/G | 1 | 0 |
| 28 | C/C | C/C | 0 | 1 |
| 29 | G/G | C/C | 1 | 0 |
| 30 | T/T | C/C | 1 | 0 |
| 31 | G/G | G/G | 0 | 1 |
| 32 | C/C | C/C | 0 | 1 |
| 33 | A/C | A/A | 0.5 | 0.5 |
| 34 | G/G | G/G | 0 | 1 |
| 35 | C/T | C/C | 0.5 | 0.5 |
| 36 | C/T | C/C | 0.5 | 0.5 |
| 37 | skip | skip | ||
| 38 | skip | skip | ||
| 39 | A/G | A/A | 0.5 | 0.5 |
| 40 | A/C | A/A | 0.5 | 0.5 |
| 41 | A/C | A/A | 0.5 | 0.5 |
| 42 | A/A | A/G | 0.5 | 0.5 |
| 43 | skip | skip | ||
| 44 | A/T | A/T | 0.5 | 0.5 |
| 45 | A/G | A/A | 0.5 | 0.5 |
| 46 | C/T | C/T | 0.5 | 0.5 |
| 47 | C/T | C/C | 0.5 | 0.5 |
| 48 | G/T | G/T | 0.5 | 0.5 |
| 49 | A/G | A/A | 0.5 | 0.5 |
| 50 | A/C | C/C | 0.5 | 0.5 |
| 51 | T/T | T/T | 0 | 1 |
| 52 | T/T | G/G | 1 | 0 |
| 53 | skip | skip | ||
| 54 | skip | skip | ||
| 55 | T/T | C/T | 0.5 | 0.5 |
| 56 | C/G | C/C | 0.5 | 0.5 |
| 57 | C/T | C/C | 0.5 | 0.5 |
| 58 | C/T | C/C | 0.5 | 0.5 |
| 59 | A/A | T/T | 1 | 0 |
| 60 | C/C | C/C | 0 | 1 |
| 61 | A/A | A/A | 0 | 1 |
| 62 | C/G | C/C | 0.5 | 0.5 |
| 63 | A/G | A/G | 0.5 | 0.5 |
| 64 | G/T | G/T | 0.5 | 0.5 |
| 65 | C/T | C/T | 0.5 | 0.5 |
| 66 | T/T | G/G | 1 | 0 |
| 67 | A/T | A/T | 0.5 | 0.5 |
| 68 | A/C | A/C | 0.5 | 0.5 |
| 69 | C/G | C/C | 0.5 | 0.5 |
| 70 | C/T | C/T | 0.5 | 0.5 |
| 71 | T/T | G/G | 1 | 0 |
| 72 | skip | skip | ||
| 73 | G/G | G/G | 0 | 1 |
| 74 | C/C | A/C | 0.5 | 0.5 |
| 75 | G/G | A/G | 0.5 | 0.5 |
| 76 | T/T | T/T | 0 | 1 |
| 77 | C/T | T/T | 0.5 | 0.5 |
| 78 | G/G | C/G | 0.5 | 0.5 |
| 79 | C/T | T/T | 0.5 | 0.5 |
| 80 | C/T | C/C | 0.5 | 0.5 |
| 81 | G/T | G/T | 0.5 | 0.5 |
| 82 | C/T | C/T | 0.5 | 0.5 |
| 83 | A/A | A/A | 0 | 1 |
| 84 | C/T | C/C | 0.5 | 0.5 |
| 85 | T/T | G/G | 1 | 0 |
| 86 | skip | skip | ||
| 87 | G/T | G/T | 0.5 | 0.5 |
| 88 | C/T | C/T | 0.5 | 0.5 |
| 89 | C/T | T/T | 0.5 | 0.5 |
| 90 | skip | skip | ||
| 91 | G/G | G/G | 0 | 1 |
| 92 | T/T | G/G | 1 | 0 |
| 93 | G/T | G/T | 0.5 | 0.5 |
| 94 | A/G | — | skip | skip |
| 95 | A/G | A/G | 0.5 | 0.5 |
