About the Registry
Welcome to the USYAKS on-line registry. This is where USYAKS tracks and stores DNA data, yak pedigrees, and yak ownership.
The USYAKS registry is a science-based registry. Yaks are admitted to or excluded from the USYAKS registry entirely based on their DNA. The North America yak gene pool is very narrow. USYAKS promotes preservation of the yak genome in North America and increased genetic diversity through selective breeding. USYAKS does not exclude yaks from the registered gene pool for non-scientific reasons.
The pedigree, the list of offspring, and the actual DNA base pairs for every yak in our registry is available for viewing and searching.
The USYAKS registry is entirely on-line. Below is a screenshot of part of HSY Becca 12D13’s Registration Certificate.
How to read and interpret Becca’s certificate as an example is described below.
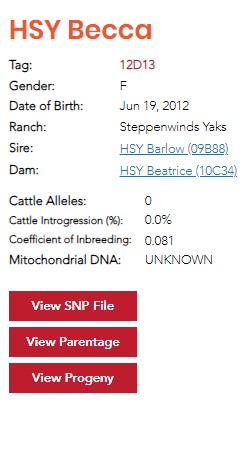
HSY Becca is the name of this yak. Becca is the given name. HSY is the herd ID, all yaks registered by Hay Springs Yaks include HSY as part of the yak’s name.
Tag: 12D13 is the UYAKS ID for HSY Becca. There can be more than one Becca, but there can only be one 12D13. The “12” indicates that Becca was born in 2012, the D13 is a computer assigned number. Most official references to Becca will refer to her as HSY Becca 12D13.
Ranch: Steppenwinds Yaks, HSY Becca is owned by Steppenwinds Yaks, one of our member ranches.
Sire and Dam: Notice that these are links, clicking on one of these will take you to the Certificate for that yak.
Cattle Alleles: The USYAKS DNA testing refers to 174 alleles. The DNA test can tell the difference between a yak allele versus an allele that originated in cattle. Yaks were domesticated by crossing them with cattle, so all yaks, even wild Tibetan yaks, have some cattle alleles. The number of cattle alleles is monitored to track yaks that were produced by recent hybridization.
Coefficient of Inbreeding (COI): Coefficient of Inbreeding has traditionally been calculated (using a relatively simple formula) by monitoring pedigrees for common ancestors. This technique does not work very well with yaks because of the narrow gene pool in North America which results in shorter pedigrees. It is impossible to calculate meaningful COI using the traditional method in instances of short pedigree. North American yaks are believed to be descended from an original population of imported yaks that might only have numbered 100 or so! With such little genetic variation present, it’s possible to have genetic commonality appearing in yaks without recent common ancestors. Therefore, the COI is calculated directly from the 174 alleles used in the genetic test in a scientific manner. A COI percentage will indicate inbreeding if there is an increased likelihood of having matched base-pairs at each DNA site. By quantifying the frequency of base pair matches we can estimate coefficient of inbreeding completely independent of pedigree and more science-based.
To interpret the number displayed in a COI result, from a mathematical standpoint, every COI number is between -1.0 and +1.0. The average COI is zero. A COI in the positive range means a yak is more in-bred than average. A COI in the negative range indicates less in-breeding than average. About 50% of registered yaks have a COI between -0.10 and +0.10. In practice, the measured COIs vary between about +0.45 (most in-bred) and -0.45 (most-outcrossed). Becca’s COI of +0.08 is pretty close to the registered herd’s average COI. She’s a bit more in-bred than the average yak.
If a yak is bred to a very close relative, the offspring’s COI will increase significantly from the dam’s and the sire’s. Just as COI can be greatly increased in one generation, it can also be greatly decreased in one generation. You can see each yak’s actual DNA base pairs by clicking on the tab <View SNP File>. The SNP data in USYAKS DNA test allows a breeder to perform virtual pairings. The COI of a calf can be predicted based on the DNA base pairs of the candidate parents; our Match-A-Yak program on the membership page will do this for you.
The DNA in a cell’s mitochondrion is inherited entirely from the mother. Tracking of mitochondrial DNA (mtDNA) allows for the tracking of a direct maternal line back to the time of species differentiation.
USYAKS DNA test is being modified to test for all four of these mtDNA varieties. Right now, it can only identify the mtDNA that originated from wild yaks of the Tibetan plateau. Becca’s mtDNA type is listed as “unknown”. Her mtDNA almost certainly came from Asian cattle species thousands of years ago during domestication. It’s possible that her mtDNA originated in Bison or from North American cattle. We hope that an improved DNA test will be able to sort this out very soon. The entire Registry is searchable, feel free to poke around.
